Knowledge has different forms such as scientific papers, raw data, and even images. In most cases, knowledge can only be fully understood if the context is clear. For example, a table with concentrations in a reactor is only useful if the user knows the reactor and the substances used. Often there is no general, formal conceptualization of the knowledge presented. As a result, the data can only be understood in the context of its presentation. An ontology is an explicit specification of a conceptualization. Thus, ontologies are used to represent relationships between concepts and provide a more detailed representation of the relationships between these concepts.
Click here for more information:
Would you like to learn how to use ontologies? 
NFDI4Cat provides you a guide with the basic steps to help you get started.
Step 1: The Domain
Define and delimit the domain of interest. This step is necessary to ensure that the ontology is well defined and the boundaries to other domains are clear.
Step 2: The Collection of Terms
Collect the main concepts to describe the defined and delimited domain. Next, sort these concepts in discussions with as many experts as possible. These discussions should also provide insight into the hierarchical structure of the concepts.
Step 3: The Search
Start by searching for existing ontologies for the concepts collected in the previous step. You can do this either manually or via an API on websites like Ontology Lookup Service or BioPortal .
Step 4: The expansion
Have you found an ontology that contains most of the concepts relevant to your domain? Expand this ontology by including the missing terms.
Step 5: Publication and Iteration
Ontology development is a collaborative process. Hence, once you are ready, you should share your ontology with other researchers in accordance with the FAIR principles. Iteration in general and at each step presented is strongly recommended until the ontology is sufficient for your purposes.
Searching the right ontology
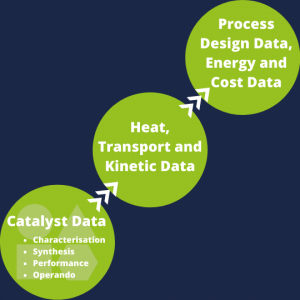
To address the problem of finding the right ontology and to facilitate the process, NFDI4Cat provides an overview of the most important ontologies in the field of catalysis research. The ontologies are grouped according to their position in the digital value chain of catalysis:
- Catalysis data (Synthesis, operando, performance, and characterisation data),
- Heat, transport, and kinetic data,
- Process design, and
- Energy and costs data.
The most relevant top-level ontologies are also available.
The ontology collection will be hosted by TIB along with the ontology collections of other NFDIs as of January 2023.
Learn MoreAFO
Allotrope Foundation Ontologies
Performance DataThe AFO is an ontology suite that provides a standard vocabulary and semantic model for the representation of laboratory analytical processes. The AFO suite is aligned at the upper layer to the Basic Formal Ontology (BFO). The core domains modeled include: Equipment, Material, Process, and Results.
BFO
Basic Formal Ontology
Top Level OntologyThe Basic Formal Ontology (BFO) is a small, upper level ontology that is designed for use in supporting information retrieval, analysis and integration in scientific and other domains. BFO is a genuine upper ontology. Thus it does not contain physical, chemical, biological or other terms which would properly fall within the coverage domains of the special sciences. BFO is used by more than 250 ontology-driven endeavors throughout the world.
CAO
Chemical Analysis Ontology
Characterisation Data Heat, Transport and Kinetic DataThe Chemical Analysis Ontology (CAO) is organized using the BFO framework with entries primarily under concepts (not complete by any means), material entities, information content entities (data items), roles, and processes. Currently, the ontology is more of a vocabulary in that development of predicates (ontology properties) that relate subjects to objects has not been a focal point.
ChEBI
Chemical methods ontology
Synthesis DataChEBI is a freely available, open source bioinformatics and cheminformatics resource that provides manually annotated information on ‘small’ chemical entities – constitutionally or isotopically distinct atoms, molecules, ion pairs, radicals, radical ions, complexes, conformers, or anything else that is a distinguishable entity. In biology, metabolites are a typical example of small molecules. Molecules directly encoded by the genome (e.g. nucleic acids, proteins and peptides derived from proteins by cleavage) are not as a rule included in ChEBI.
CHEMINF
Chemical Information Ontology
Characterisation DataThe Chemical Information Ontology (CHEMINF) aims to establish a standard in representing chemical information. In particular, it aims to produce an ontology to represent chemical structure and to richly describe chemical properties, whether intrinsic or computed.
CHMO
Chemical methods ontology
Synthesis Data Operando Data Performance DataCHMO, the chemical methods ontology, describes methods used to collect data in chemical experiments, such as mass spectrometry and electron microscopy prepare and separate material for further analysis, such as sample ionisation, chromatography, and electrophoresis synthesise materials, such as epitaxy and continuous vapour deposition It also describes the instruments used in these experiments, such as mass spectrometers and chromatography columns. It is intended to be complementary to the Ontology for Biomedical Investigations (OBI).
CIF-Ontology
Crystallography domain ontology
Characterisation DataA crystallography domain ontology based on EMMO and the CIF core dictionary. It is implemented as a formal language.
DOLCE
Descriptive Ontology for Linguistic and Cognitive Engineering
Top Level OntologyDOLCE is a foundational ontology developed and maintained by the ISTC-CNR Laboratory for Applied Ontology . It was originally developed within the WonderWeb project and was conceived as the first module of the WonderWeb Foundational Ontologies Library (WFOL) together with OCHRE and BFO. DOLCE has remained stable after its first release in 2002/2003. It is formally specified in first-order logic (FOL).
EMMO
European Materials & Modelling Ontology
Top Level OntologyEMMO is a multidisciplinary effort to develop a standard representational framework (the ontology) for applied sciences. It is based on physics, analytical philosophy and information and communication technologies. It has been instigated by materials science and provides the connection between the physical world, the experimental world (materials characterisation) and the simulation world (materials modelling). It is released under a Creative Commons CC BY 4.0 license.
ISO 15926
ISO 15926 interoperability standard
Heat, Transport and Kinetic Data Process Design Energy and Cost DataThe full title of ISO15926 is “Industrial Automation Systems and Integration of Life-cycle Data for Process Plants, including Oil and Gas Production Facilities.” It is about the standardization of the flow of information. The word “Integration” in the title means that by using ISO 15926 all applications can exchange information with each other without having to modify the applications in any way.
ISO 15926-14
ISO 15926 Part 14: Data model adapted for OWL2 direct semantics
Top Level OntologyThe primary purpose of ISO 15926 is to provide a foundation ontology to support the integration and sharing of data related to the lifecycle of a process plant in such a way that it is consistent, unambiguous, and minimizing the number of ways something could be expressed. Further purpose of ISO 15926-14 is to meet needs for OWL 2 ontologies that are based on ISO 15926-2, that enable efficient reasoning and that capture lifecycle information. A specific purpose is to demonstrate lifecycle modelling through a representation of the lifecycle model of ISO/IEC 81346-1. Another specific purpose is to exemplify how this standard can be used to develop industrial ontologies through various real-world use cases from industy.
OntoCAPE
Ontology for computer aided process engineering
Heat, Transport and Kinetic Data Process Design Energy and Cost DataOntoCAPE is a large-scale ontology for the domain of Computer Aided Process Engineering (CAPE). Represented in a formal, machine-interpretable ontology language, OntoCAPE captures consensual knowledge of the process engineering domain in a generic way such that it can be reused and shared by groups of people and across software systems. On the basis of OntoCAPE, novel software support for various engineering activities can be developed; possible applications include the systematic management and retrieval of simulation models and design documents, electronic procurement of plant equipment, mathematical modeling, as well as the integration of design data from distributed sources.
OntoCompChem
Computational Chemistry Ontology
Synthesis Data Operando DataLinked-data framework for connecting species in chemical kinetic reaction mechanisms with quantum calculations. A mechanism can be constructed from thermodynamic, reaction rate, and transport data that has been obtained either experimentally, computationally, or by a combination of both. In order to implement this approach, two existing ontologies, namely OntoKin, for representing chemical kinetic reaction mechanisms, and OntoCompChem, for representing quantum chemistry calculations, are extended.
OntoKin
Ontology for Chemical Kinetic Reaction Mechanisms
Operando Data Performance DataAn ontology for capturing both data and the semantics of chemical kinetic reaction mechanisms has been developed. Such mechanisms can be applied to simulate and understand the behavior of chemical processes, for example, the emission of pollutants from internal combustion engines. An ontology development methodology was used to produce the semantic model of the mechanisms, and a tool was developed to automate the assertion process. As part of the development methodology, the ontology is formally represented using a web ontology language (OWL), assessed by domain experts, and validated by applying a reasoning tool. The resulting ontology, termed OntoKin, has been used to represent example mechanisms from the literature. OntoKin and its instantiations are integrated to create a knowledge base (KB), which is deployed using the RDF4J triple store. The use of the OntoKin ontology and the KB is demonstrated for three use cases—querying across mechanisms, modeling atmospheric pollution dispersion, and as a mechanism browser tool. As part of the query use case, the OntoKin tools have been applied by a chemist to identify variations in the rate of a prompt NOx formation reaction in the combustion of ammonia as represented by four mechanisms in the literature. Merged with OntoCAPE.
OSMO
Ontology for simulation, modelling, and optimization
Operando Data Heat, Transport and Kinetic Data Process Design Energy and Cost DataOSMO is an ontologization and extension of MODA, a workflow metadata standard that constitutes a mandatory requirement within a number of European calls and projects in the context of materials modelling. OSMO was developed within the Horizon 2020 project VIMMP (Virtual Materials Marketplace) and is part of a larger effort in ontology engineering driven by the European Materials Modelling Council, with the European Materials and Modelling Ontology (EMMO) as its core.
REX
REX
Performance Data Characterisation DataAn ontology of physico-chemical processes, i.e. physico-chemical changes occurring in course of time. This ontology is not well structured and lacks of properties. Also the design principle is not clear. Nevertheless it provides a good amount of concepts important to macroscopic and microscopic physical processes in form of a more hierarchical structure.
RXNO
Name Reaction Ontology
Synthesis DataRXNO is the name reaction ontology. It contains more than 500 classes representing organic name reactions such as the Diels–Alder cyclization and the Cannizzaro reaction to their roles in an organic synthesis.
SBO
Systems Biology Ontology
Operando Data Performance Data Characterisation DataThe Systems Biology Ontology is a set of controlled, relational vocabularies of terms commonly used in Systems Biology, and in particular in computational modelling. It consists of seven orthogonal vocabularies defining: reaction participants roles (e.g. “substrate”), quantitative parameters (e.g. “Michaelis constant”), classification of mathematical expressions describing the system (e.g. “mass action rate law”), modelling framework used (e.g. “logical framework”), the nature of the entity (e.g. “macromolecule”), the type of interaction (e.g. “process”), as well as a branch to define the different types of metadata that may be present within a model.
Would you like to do more than stay informed?
You can join NFDI4Cat as a member and actively shape the digital future of catalysis!
Contact us